Structural variants (SV) are recognized as a prominent source of genetic variation, with single variants manifesting significant influence over phenotypic expression. However, their effects remain poorly understood due to computational limitations and a continued focus on SNPs. Addressing this need, we introduce the Variant Identification Pipeline with Expression Realization (VIPER), a robust computational pipeline created to identify and annotate structural variants with significant impact on gene expression. Using a publicly available structural variant collection as reference (Alonge et al., 2020), VIPER accepts processed RNA sequence data and constructs a list of SV-gene pairs based on genetic position. These SV-gene pairs are then evaluated by their differential expression values, with pairs expressing significant RPKM differentials concatenated to form a library of high-impact SV-gene pairs. Finally, the library is enriched by annotating genes for function, allowing for the identification and selection of candidate variants with desirable phenotypic influences.
To evaluate the efficacy of VIPER, we constructed a causal genomic library from a collection of 108 tomato leaf samples. A candidate insertion from this library, which we labelled GUNGIR, affirmed VIPER by expressing a significant influence over leaflet morphology as quantified by a fivefold principal component system.
RNA Data Preparation
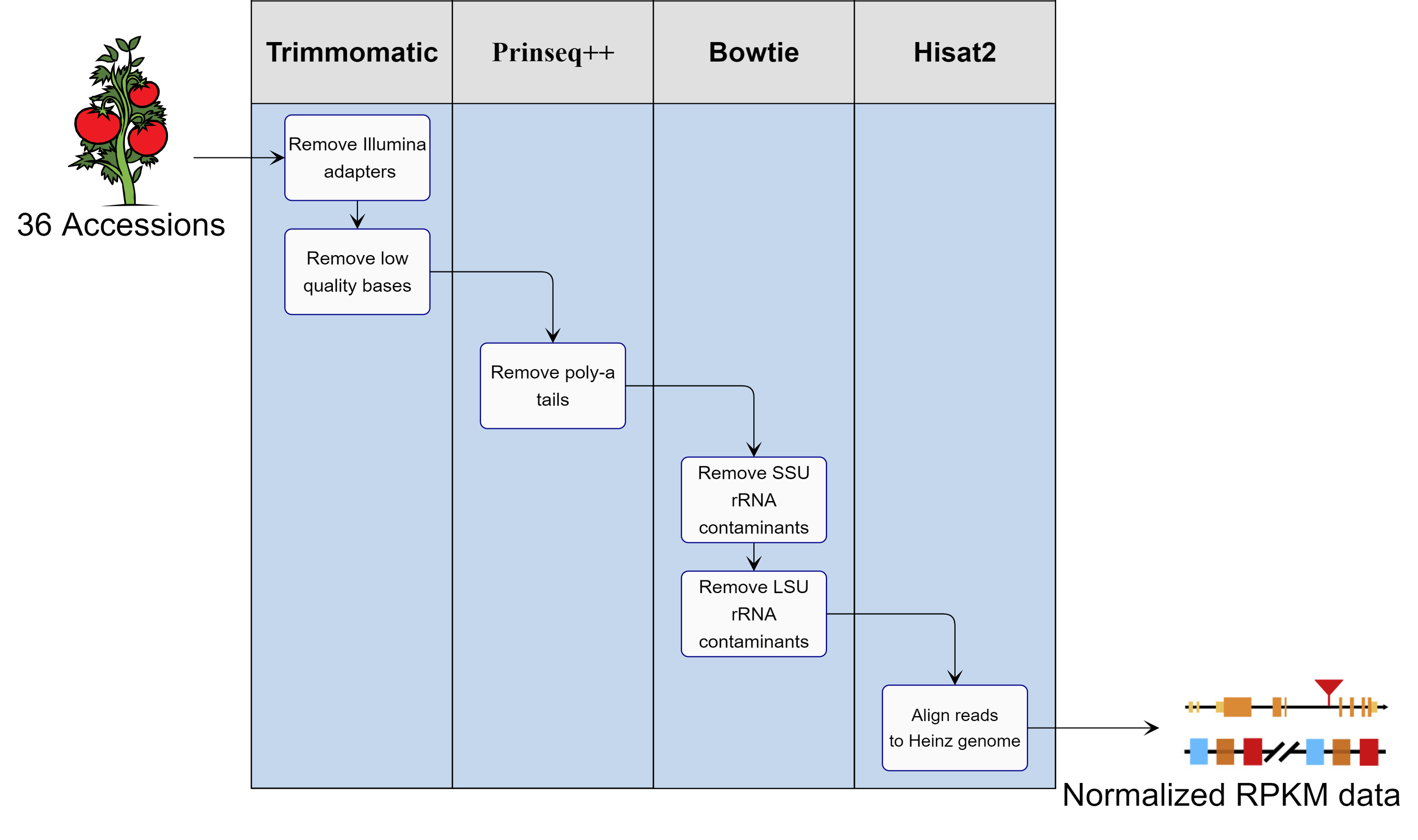
Figure 1. A visualization of the pipeline created to process our raw RNA sequence data. The data originated from the leaf material of 36 tomato accessions with 3 biological replicates per accession, culminating in 108 total samples. Each sample was trimmed for various contaminants and subsequently aligned with the Heinz tomato genome to generate RPKM data. The RPKM values were normalized and merged by accession using homebrew scripts in conjunction with the Fei lab to generate single RPKM values for each accession.
SV-Gene Identification
- RNA-sequence processing provided normalized RPKM values for 34,693 total genes.
- When cross referenced with the publicly available PanSV structural variant library, 5,471 genes overlapped with a structural variant, allowing for genic SV-gene pairing. (Figure 3)
- 3,543 of the genic pairs met the minimum expression requirement by expressing in at least five accessions, and were statistically ranked by evaluating differential expression across an accession split. (Figure 4)
- For each of the 3,542 evaluated accessions splits, the 34,692 non-paired genes were similarly evaluated for differential expression. (Figure 5)
- This quantitative evaluation constructed a library of 271 statistically significant SV-gene pairs.
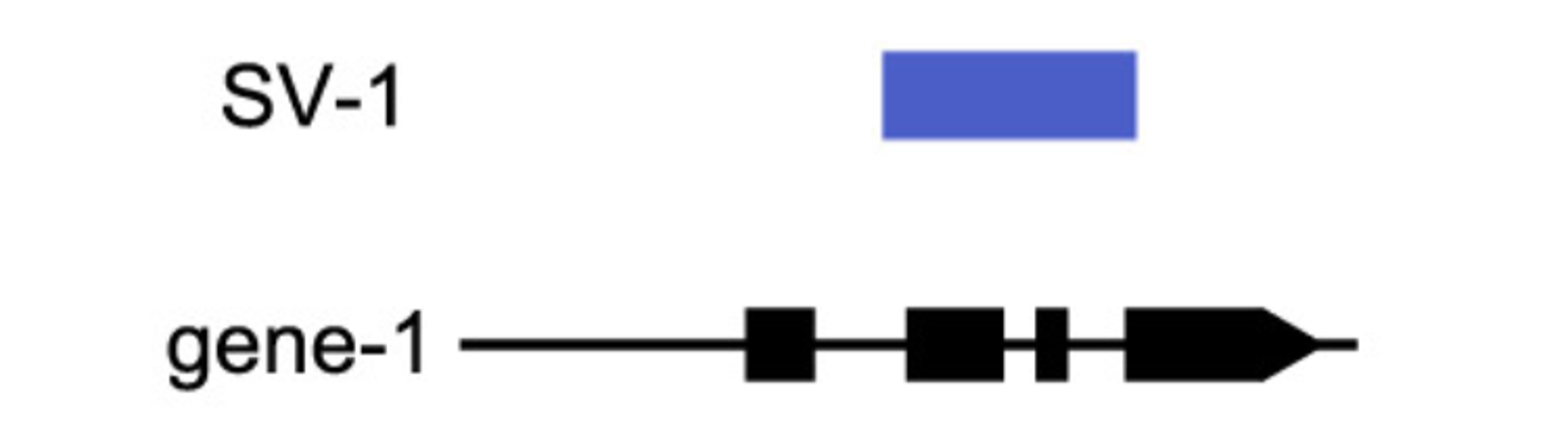
Figure 2. A visualization of what qualifies as an SV-gene pair (modified after Alonge et al., 2020). We focused on genic structural variants as potential pairs to test our pipeline, disregarding variants in both upstream and downstream regions. Future work will include consideration of adjacent non-coding sequence SVs.
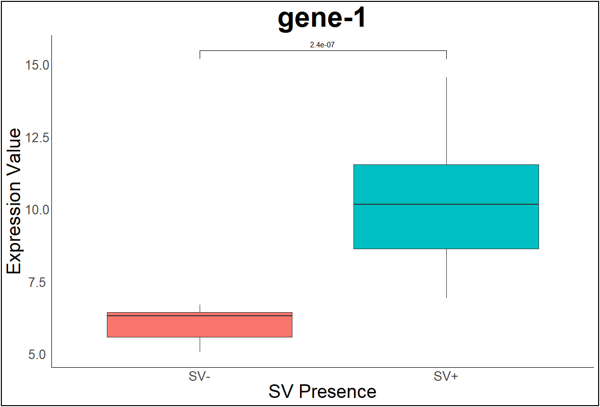
Figure 3. The significant difference in expression between accessions with the SV-gene pair and those without in a high-impact structural variant. Each of the 3,543 genic SV-gene pairings underwent this accession split to quantify their expression. E.g. gene-1: Solyc01g005100.
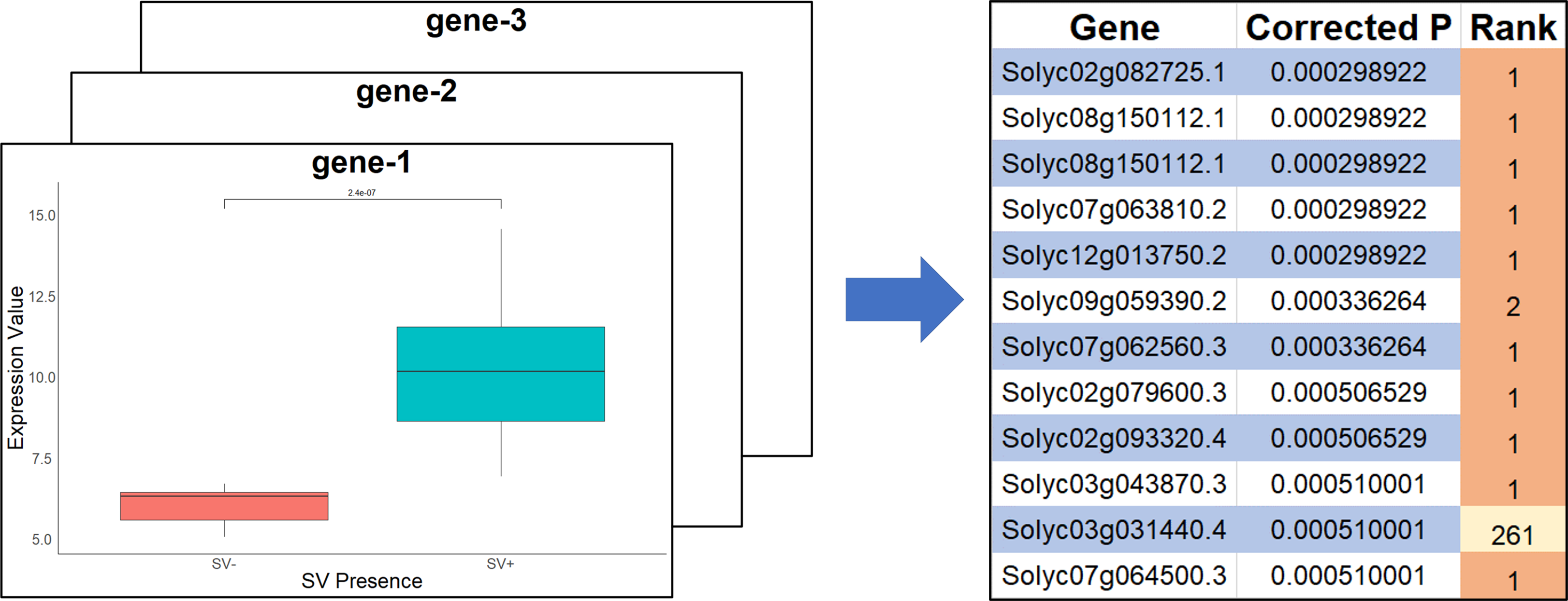
Figure 4. For each accession split, differential expression was also calculated for each of the 34,692 non-paired genes. This allowed for the identification of genes whose expression was impacted by SV presence/absence, but were not identified by position. E.g. genes: Solyc01g005100, Solyc01g005430.3, Solyc01g005430.3.
Single Variant Influence
- To evaluate the functional efficacy of our 271 high-impact SV-gene pairs, we identified 103 overlapping genes between our 271 SV-gene pairs and Martinez, 2021, a set of literature-curated genes related to leaf development.
- Differences in the leaf development of our tomato accessions were quantified by the Sinha Lab at UC Davis, by means of a fivefold principal component system. (Figure 7)
- The highest-ranking of the isolated SV-gene pairs, a 143 bp insertion we labelled GUNGIR, expressed a strong phenotypic influence over leaflet shape in our tomato accessions. GUNGIR localizes in the candidate tomato gene Solyc01g005100, encoding a 2-oxoglutarate and Fe(II)-dependent oxygenase superfamily protein.
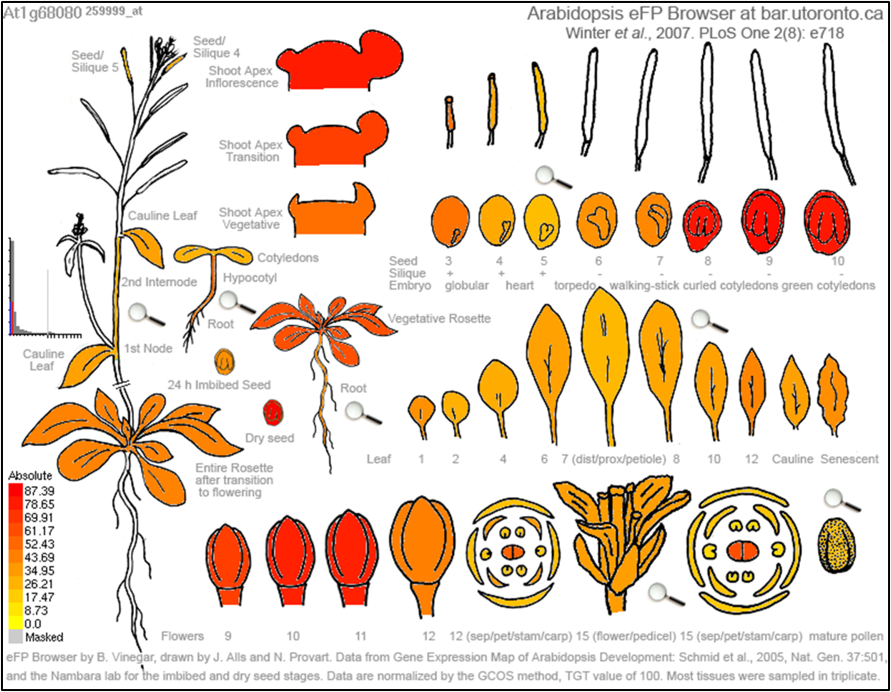
Figure 5. An expression atlas for the Arabidopsis homolog of GUNGIR. Areas of high expression include early flower development and late shoot and seed iterations.
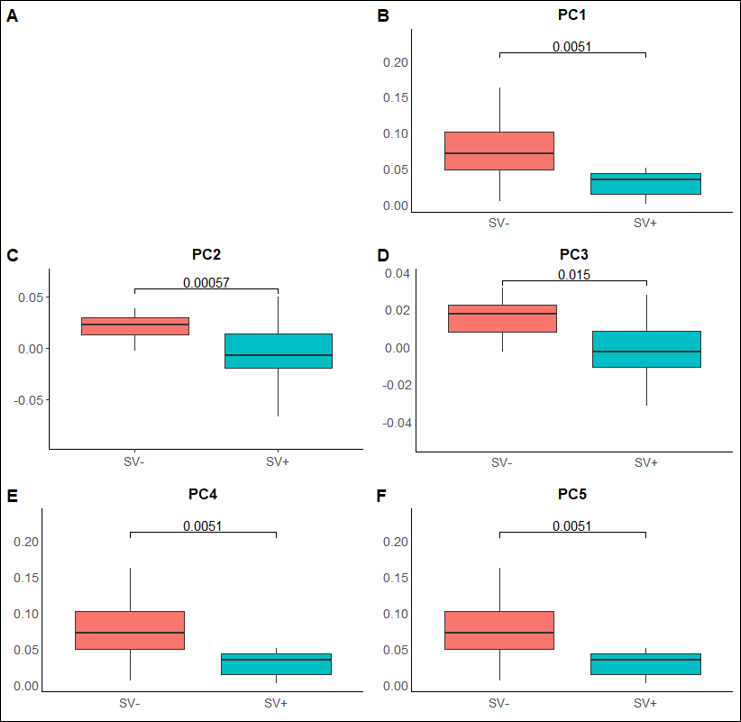
Figure 6. Morphological analysis of the leaflets of our tomato accessions by the Sinha Lab at UC Davis generated five principal components, correlated to traditional shape measures, with which to quantify leaf shape. In an accession split based on GUNGIR presence/absence, the leaflets expressed significant differences in all five principal components.
Phenotypic Impact

Figure 7. Leaflets photographed from four accessions that do not possess GUNGIR. Leaflets trended towards a more uniform aspect ratio and less serrated edge.

Figure 8. Leaflets photographed from four accessions that do possess GUNGIR. Leaflets trended towards a less uniform aspect ratio with a reduced Tip:Base differential and an increased serration and lobing density.