Senior Legacy Presentation
Tracking Antigen Exposure Using Deep Neural Networks
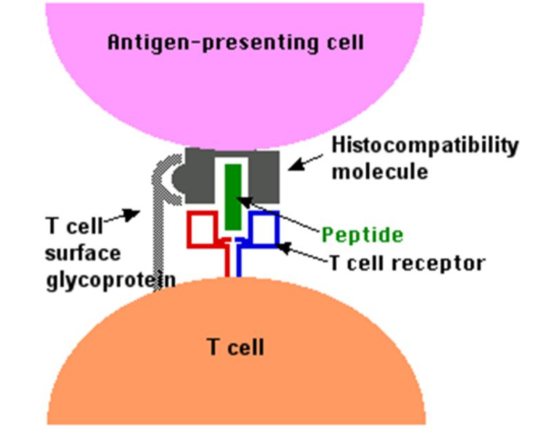
Because infection causes expansion of pathogen specific T cells that express pathogen specific unique receptors encoded in DNA, exposure history of an individual can be accessed through examination of their T-cell receptors (TCRs). These unique sequences can be used as biomarkers for tracking T-cell responses and cataloging immunological history. The...
[Read More]